最近想要可视化样本间的相关性,但又不满足于常规的相关性热图。因此,就注意到GGally包中的ggpairs函数,可以方便地实现多方面的相关性可视化。
本文仅介绍
ggpairs在连续型变量方面的应用。它也可以用到离散型变量的可视化上。
下面以airway数据集进行演示:
这里我们在前4个样本中随机选取1000个基因进行展示
1 | library(GGally) |
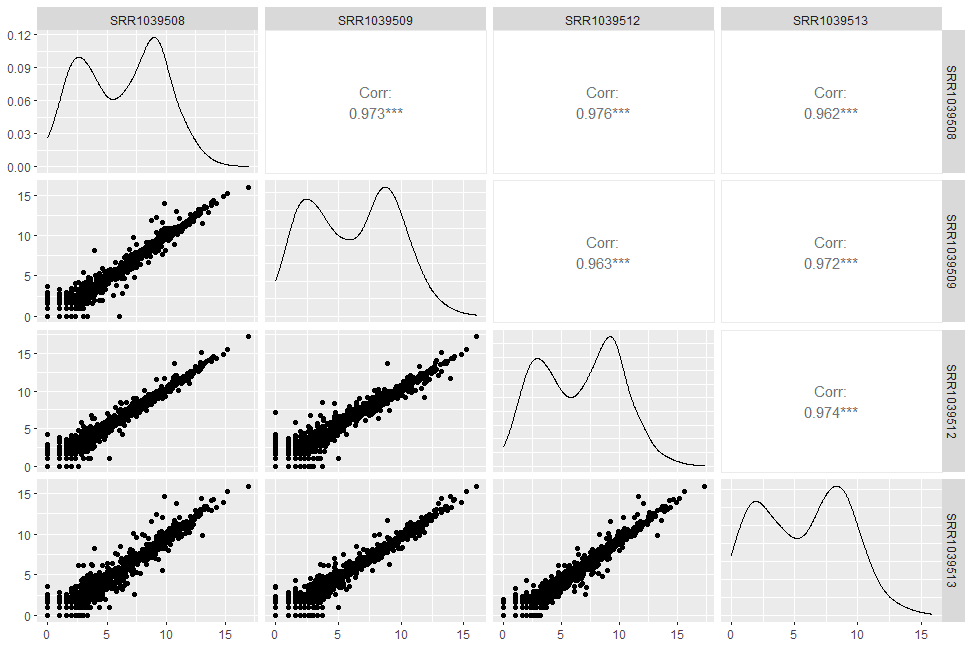
ggpairs将输出的图划分为三个区域,分别是左下角的lower, 对角线的diag, 以及右上角的upper. 对于连续性数值变量,默认在lower区画pairwise scatter plot,diag区画density plot,upper区展示相应的pairwise Pearson’s correaltion coefficient.
进一步,我还希望在左下角的散点图中加入y=x的拟合线,并在对角线的加上直方图。我们可以通过自定义画图的函数实现这些操作。
1 | ggscatter <- function(data, mapping, ...) { |
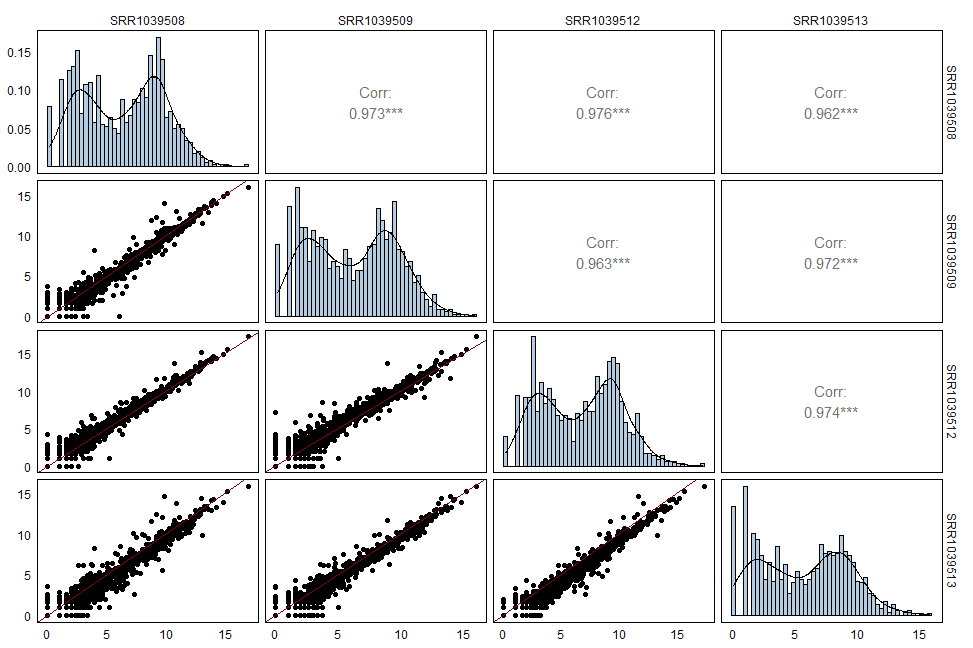
再放一个高度修改的版本
1 | # https://pascal-martin.netlify.app/post/nicer-scatterplot-in-gggally/ |
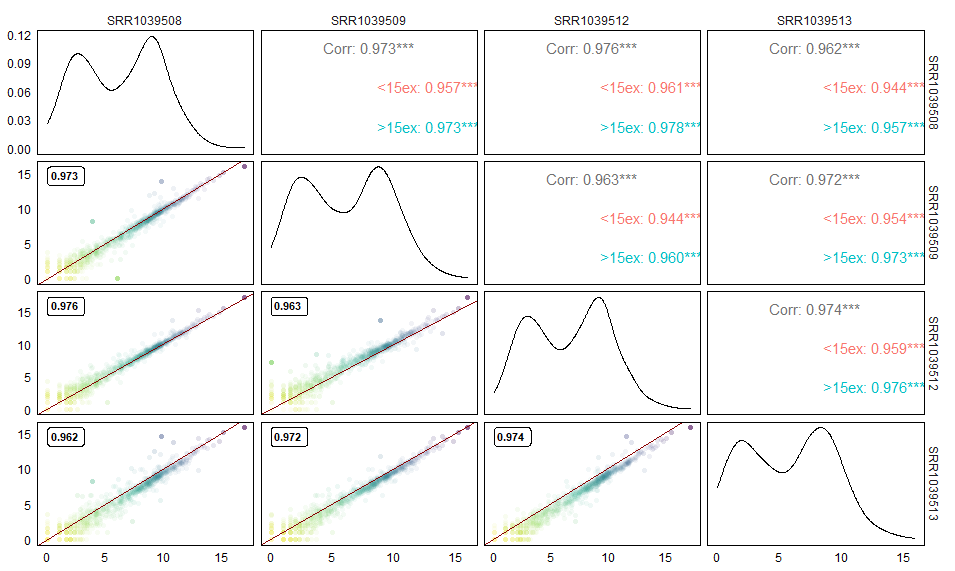
- 对散点图的透明度进行调整,点越密的区域透明度越高,反之亦然。
- 散点图的颜色代表了基因表达量。
- 在散点图左上角添加Pearson’s 相关系数
- 右上角展示了对小于15个外显子的基因和大于15个外显子基因的相关性的统计。
在我看来ggpairs相当于是一个ggplot2的集成可视化方法,可以很方便的一次性展示多个方面的相关性信息。同时,它的可定制性也很高,可以满足许多额外的可视化需求。唯一的缺陷可能是需要耗费一定功夫写出包装的函数。
Ref:
ggpairs doc: https://ggobi.github.io/ggally/articles/ggpairs.html
Nicer scatter plots in GGgally ggpairs-ggduo: https://pascal-martin.netlify.app/post/nicer-scatterplot-in-gggally
Creating a density histogram in ggplot2: https://stackoverflow.com/questions/21061653/creating-a-density-histogram-in-ggplot2
完。