本文简单介绍几种重抽样方法 (in R)。
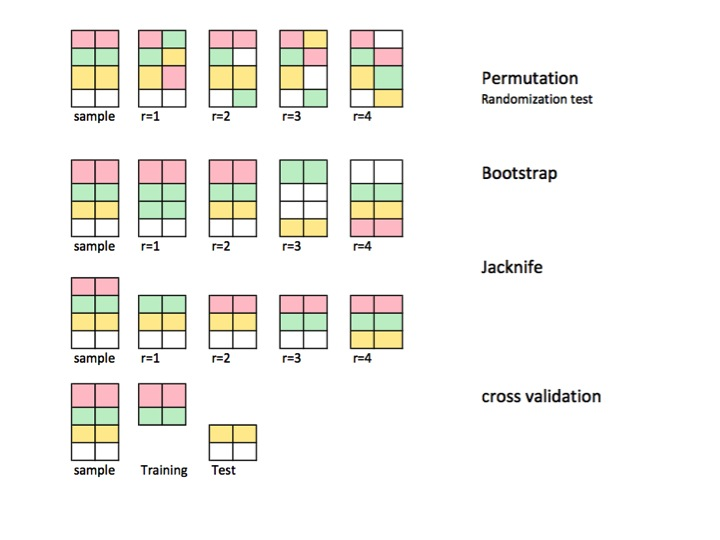
我们生成一组数据,其中x是我们的观测值,y是我们对其的标签。
1
2
3
4
5
6
|
set.seed(1111)
x <- c(rnorm(10), rnorm(10, mean=5, sd=5))
y <- c(rep("A", 10), rep("B", 10))
df1 <- data.frame(x,y)
str(df1)
|
Permutation
Permutation相当于是一种无放回的重抽样方法,通常用于假设检验。
1
2
3
|
set.seed(2222)
sample(df1$x, replace = FALSE)
|
我们可以使用Permutation test检验A,B两组的值是否有差异
1
2
3
4
5
6
7
8
9
10
11
12
13
|
set.seed(3333)
permt.ls <- list()
for (i in 1:1000) {
permt.i <- sample(df1$x, replace = FALSE)
diff.i <- abs(mean(permt.i[1:10]) - mean(permt.i[11:20]))
permt.ls[[i]] <- c(permt.i, diff.i)
}
permt.df <- Reduce(rbind, permt.ls)
diff.raw <- abs(mean(df1$x[1:10]) - mean(df1$x[11:20]))
mean(permt.df[,21] >= diff.raw)
|
每一次重抽样后,我们都可以计算两组样本均值的差异。如果重抽样样本组间差异大于原始样本组间差异的话,可以认为是一次错误事件,通过计算错误事件在总重抽样次数中的占比就可以得到置换检验的p值。
在这里1000次重抽样中,只有79次是错误事件,所以我们的p值为0.079
Bootstrap
Bootstrap是一种有放回的重抽样方法,通常用于参数估计。
1
2
3
|
set.seed(4444)
sample(df1$x, replace = TRUE)
|
例如,我们用bootstrap估计总体的均值
1
2
3
4
5
6
7
8
9
10
| set.seed(5555)
mean.raw <- mean(df1$x)
mean.i <- c()
for (i in 1:1000) {
boot.i <- sample(df1$x, replace = TRUE)
mean.i[i] <- mean(boot.i)
}
mean.boost <- mean(mean.i)
mean.raw; mean.boost
|
此外,我们还可以计算bootstrap预测均值的标准误(SE)
1
2
3
|
mean.se.boost <- sqrt(sum((mean.i - mean.boost)^2)/(1000-1))
mean.se.boost
|
Jackknife
Jakknife可以被认为是一种leave-one-out的重抽样方法,对于大小为k的数据集,将产生k个大小为k-1的样本.
1
2
3
4
5
6
7
| jack.ls <- list()
for (i in 1:nrow(df1)) {
jack.ls[[i]] <- df1[-i,]
}
length(jack.ls); dim(jack.ls[[1]])
|
Cross validation
交叉验证将数据切分为测试集和验证集,常在模型拟合中使用。例如k-fold cross validation将数据划分为k组不重叠的数据集。
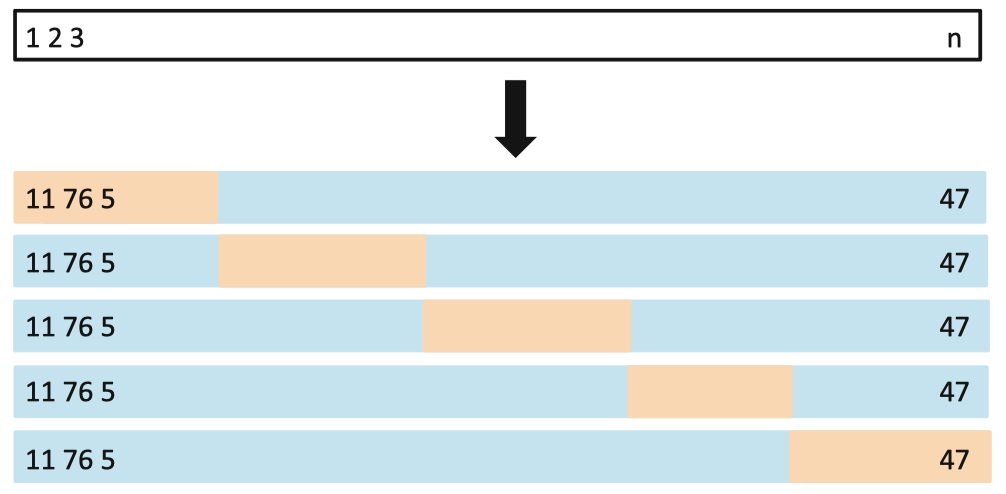
Figure: Illustration of 5-fold CV (https://yey.world/2020/08/31/MAST90083-05/).
1
2
3
4
5
6
7
8
|
set.seed(6666)
training_size <- round(nrow(df1)*0.7)
training_idx <- sample(nrow(df1), size = training_size, replace = FALSE)
training_set <- df1[training_idx,]
val_set <- df1[-training_idx,]
nrow(training_set); nrow(val_set)
|
有时候,由于样本量太小,无法满足k-fold CV中不重叠分组的要求,有的样本不可避免地被重复使用。
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
|
set.seed(7777)
fold_idx <- list()
val_size <- nrow(df1) - training_size
all_idx <- 1:nrow(df1)
for (i in 1:5) {
fold_idx_i <- unique(unlist(fold_idx))
if (all(all_idx %in% fold_idx_i) | is.null(fold_idx_i)) {
fold_idx[[i]] <- sample(all_idx, val_size, replace = FALSE)
} else if (val_size < length(all_idx[-fold_idx_i])) {
fold_idx[[i]] <- sample(all_idx[-fold_idx_i], val_size, replace = FALSE)
} else if (val_size > length(all_idx[-fold_idx_i])) {
fold_idx[[i]] <- c(all_idx[-fold_idx_i], sample(all_idx, val_size-length(all_idx[-fold_idx_i]), replace = FALSE))
}
}
fold_data <- lapply(fold_idx, function(i) {
list(training = df1[-i,], valdation = df1[i,])
})
fold_idx
|
以上就是对重抽样方法的简单介绍。
Ref:
https://stats.stackexchange.com/questions/104040/resampling-simulation-methods-monte-carlo-bootstrapping-jackknifing-cross
https://yey.world/2020/08/31/MAST90083-05/

